Functions & Debugging
Functions
Data (and objects more generally) are one of the building blocks of R. The other is functions.
Functions
Data (and objects more generally) are one of the building blocks of R. The other is functions.
We've already used a handful of functions, including seq(), arithmetic functions (+, *, etc.), c(), list(), data.frame(), str(), etc.
Functions
Data (and objects more generally) are one of the building blocks of R. The other is functions.
We've already used a handful of functions, including seq(), arithmetic functions (+, *, etc.), c(), list(), data.frame(), str(), etc.
Functions take some form of an input, perform some operation, and then return some object(s) as output.
Functions
Data (and objects more generally) are one of the building blocks of R. The other is functions.
We've already used a handful of functions, including seq(), arithmetic functions (+, *, etc.), c(), list(), data.frame(), str(), etc.
Functions take some form of an input, perform some operation, and then return some object(s) as output.
Functions are made up of arguments.
Functions
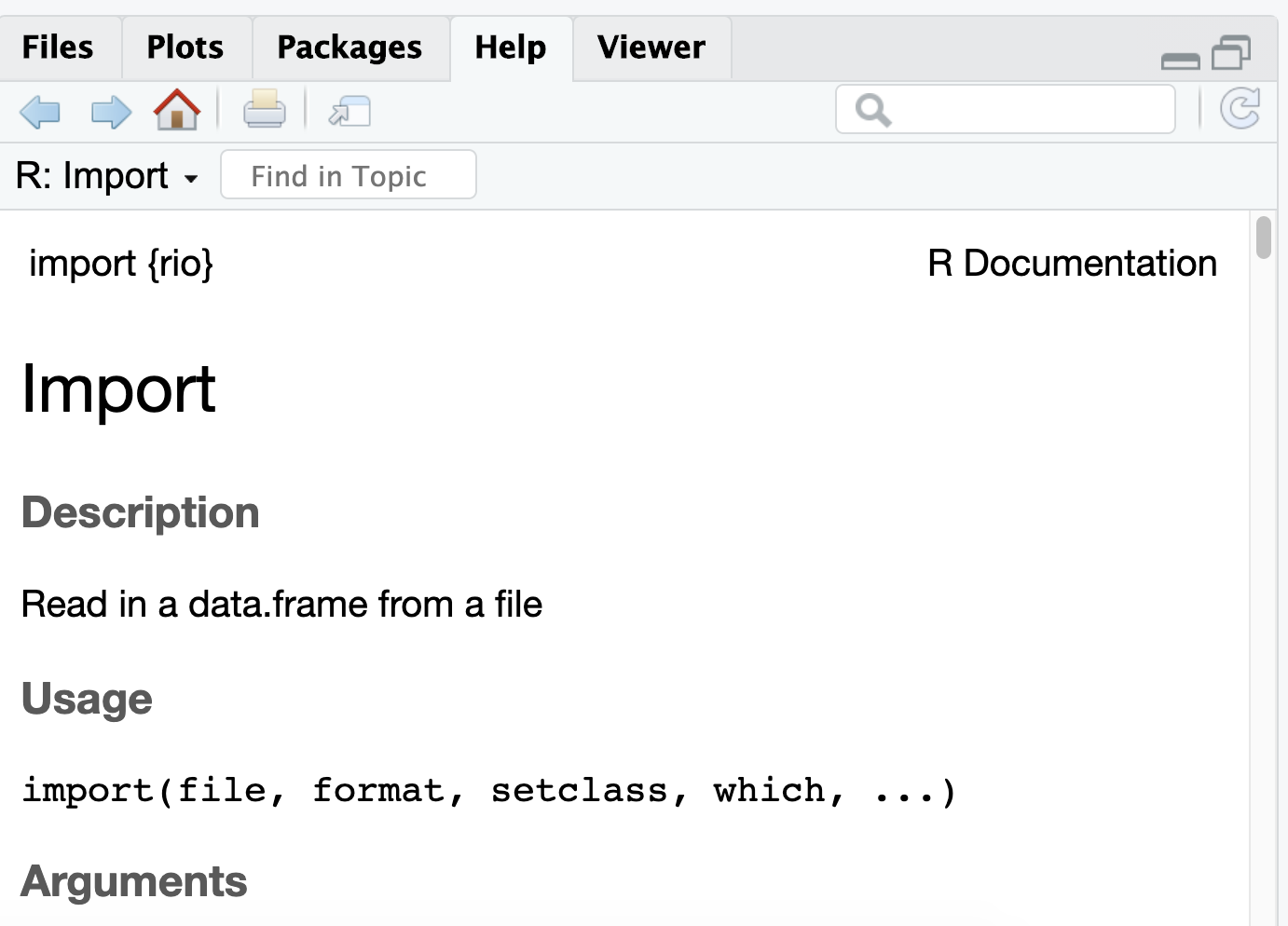
Let's take another look at the help documentation for seq()...
?seqFunctions
Let's take another look at the help documentation for seq()...
?seqYou can see it has the arguments from, to, by, length.out, and along.with.
Functions
Let's take another look at the help documentation for seq()...
?seqYou can see it has the arguments from, to, by, length.out, and along.with.
You might also notice that each of the arguments have a value after the = in the documentation.
Functions
Let's take another look at the help documentation for seq()...
?seqYou can see it has the arguments from, to, by, length.out, and along.with.
You might also notice that each of the arguments have a value after the = in the documentation.
These values are the defaults; they are what the arguments will be set to if you don't specify them.
Functions
Let's take another look at the help documentation for seq()...
?seqYou can see it has the arguments from, to, by, length.out, and along.with.
You might also notice that each of the arguments have a value after the = in the documentation.
These values are the defaults; they are what the arguments will be set to if you don't specify them.
In fact, since all of the arguments have defaults, we don't have to specify any to run seq() as we saw earlier.
seq()## [1] 1Functions
Let's take a look at a new function, mean()...
?meanFunctions
What happens if we run mean() without any arguments?
mean()## Error in mean.default(): argument "x" is missing, with no defaultFunctions
What happens if we run mean() without any arguments?
mean()## Error in mean.default(): argument "x" is missing, with no defaultWe get an error telling us that the argument "x" is missing and has no default.
Functions
What happens if we run mean() without any arguments?
mean()## Error in mean.default(): argument "x" is missing, with no defaultWe get an error telling us that the argument "x" is missing and has no default.
Whenever you see this error, it means you are missing a required argument (i.e., an argument without a default).
Functions
What happens if we run mean() without any arguments?
mean()## Error in mean.default(): argument "x" is missing, with no defaultWe get an error telling us that the argument "x" is missing and has no default.
Whenever you see this error, it means you are missing a required argument (i.e., an argument without a default).
If we look at the help documentation, you can see x is the data from which to calculate a mean.
Functions
Let's create some data to calculate the mean of.
vec <- c(1, 2, 3, 4, 5, 6, 2, 4)Functions
Let's create some data to calculate the mean of.
vec <- c(1, 2, 3, 4, 5, 6, 2, 4)Now let's take the mean of vec.
mean(x = vec)## [1] 3.375Functions
Let's create some data to calculate the mean of.
vec <- c(1, 2, 3, 4, 5, 6, 2, 4)Now let's take the mean of vec.
mean(x = vec)## [1] 3.375Note that mean() has two more optional arguments listed:
trim, which returns a trimmed meanna.rm, which takes a logical value indicating if it should remove missing values or not before it calculates the mean (FALSEby default).
Functions
What happens if we don't remove NAs before calculating the mean? Let's check it out...
Functions
What happens if we don't remove NAs before calculating the mean? Let's check it out...
vec_na <- c(1, 2, 3, 4, 5, 6, NA, 2, 4)Functions
What happens if we don't remove NAs before calculating the mean? Let's check it out...
vec_na <- c(1, 2, 3, 4, 5, 6, NA, 2, 4)mean(vec_na)## [1] NAFunctions
What happens if we don't remove NAs before calculating the mean? Let's check it out...
vec_na <- c(1, 2, 3, 4, 5, 6, NA, 2, 4)mean(vec_na)## [1] NAIt returns NA. NAs are contagious! A single NA in a vector will cause many functions to return NA (unless they remove them by default).
Functions
What happens if we don't remove NAs before calculating the mean? Let's check it out...
vec_na <- c(1, 2, 3, 4, 5, 6, NA, 2, 4)mean(vec_na)## [1] NAIt returns NA. NAs are contagious! A single NA in a vector will cause many functions to return NA (unless they remove them by default).
This sort of makes sense - the mean of vec_na in its entirety is unknown, since we don't know what the NA value is. That's why you have to remove NA's before running calculations by setting na.rm = TRUE
Functions
What happens if we don't remove NAs before calculating the mean? Let's check it out...
vec_na <- c(1, 2, 3, 4, 5, 6, NA, 2, 4)mean(vec_na)## [1] NAIt returns NA. NAs are contagious! A single NA in a vector will cause many functions to return NA (unless they remove them by default).
This sort of makes sense - the mean of vec_na in its entirety is unknown, since we don't know what the NA value is. That's why you have to remove NA's before running calculations by setting na.rm = TRUE
mean(vec_na, na.rm = TRUE)## [1] 3.375Your turn 1
02:00
Look up the help documentation for the function
sd()(type directly in the RStudio console)Calculate the standard deviation of
vec_na. Be sure to remove missing values first.
vec_na <- c(1, 2, 3, 4, 5, 6, NA, 2, 4)Functions
You can get the length of many objects with length()
length(vec_na)## [1] 9Functions
You can get the length of many objects with length()
length(vec_na)## [1] 9nrow() and ncol() can be used to get the number of rows or columns in a matrix or data frame. Let's look at the data frame df below
## a b c d## 1 1 3 5 7## 2 2 4 6 8nrow(df)## [1] 2ncol(df)## [1] 4The length of a data frame is the same as the number of columns.
length(df)## [1] 4Functions
Take another look at the help documentation for sd() 👀.
Notice that there are two arguments and they are in order, x followed by na.rm = FALSE.
Functions
Take another look at the help documentation for sd() 👀.
Notice that there are two arguments and they are in order, x followed by na.rm = FALSE.
You can set arguments explicitly by name
sd(x = vec_na, na.rm = TRUE)## [1] 1.685018Functions
Take another look at the help documentation for sd() 👀.
Notice that there are two arguments and they are in order, x followed by na.rm = FALSE.
You can set arguments explicitly by name
sd(x = vec_na, na.rm = TRUE)## [1] 1.685018You can also set them positionally and drop the argument names
sd(vec_na, TRUE)## [1] 1.685018Functions
When using arguments positionally (without their names), make sure the arguments are in the right order.
Functions
When using arguments positionally (without their names), make sure the arguments are in the right order.
Otherwise you can end up with weird errors or warnings.
sd(TRUE, vec_na)## Warning in if (na.rm) "na.or.complete" else "everything": the condition has## length > 1 and only the first element will be used## [1] NAFunctions
When using arguments positionally (without their names), make sure the arguments are in the right order.
Otherwise you can end up with weird errors or warnings.
sd(TRUE, vec_na)## Warning in if (na.rm) "na.or.complete" else "everything": the condition has## length > 1 and only the first element will be used## [1] NAHowever, if you explicitly name the arguments, you can actually put them in a different order. This isn't recommended unless there is a good reason though...
sd(na.rm = TRUE, x = vec_na)## [1] 1.685018Packages
So far, we've been working with functions that are already installed and loaded when we open R.
Packages
So far, we've been working with functions that are already installed and loaded when we open R.
However, many of the functions we want to use are not part of the basic R install. They come in packages that other R users create and share.
Packages
So far, we've been working with functions that are already installed and loaded when we open R.
However, many of the functions we want to use are not part of the basic R install. They come in packages that other R users create and share.
Most packages can be accessed from CRAN - the Comprehensive R Archive Network.
Packages
The most common way to get a package is to download it from CRAN using install.packages("package_name") -- notice the quotes.
Packages
The most common way to get a package is to download it from CRAN using install.packages("package_name") -- notice the quotes.
For example, one package we're going to use tomorrow is rio, which has really easy functions for importing and exporting data.
If we wanted to install the rio package, we would use
install.packages("rio")Packages
The most common way to get a package is to download it from CRAN using install.packages("package_name") -- notice the quotes.
For example, one package we're going to use tomorrow is rio, which has really easy functions for importing and exporting data.
If we wanted to install the rio package, we would use
install.packages("rio")A couple notes here.
1) You will sometimes see package names written inside {}, e.g. {rio}.
Packages
The most common way to get a package is to download it from CRAN using install.packages("package_name") -- notice the quotes.
For example, one package we're going to use tomorrow is rio, which has really easy functions for importing and exporting data.
If we wanted to install the rio package, we would use
install.packages("rio")A couple notes here.
1) You will sometimes see package names written inside {}, e.g. {rio}.
2) To make things easier in our online format, I have pre-installed all the packages we will be needing on RStudio Cloud.
Packages
The most common way to get a package is to download it from CRAN using install.packages("package_name") -- notice the quotes.
For example, one package we're going to use tomorrow is rio, which has really easy functions for importing and exporting data.
If we wanted to install the rio package, we would use
install.packages("rio")A couple notes here.
1) You will sometimes see package names written inside {}, e.g. {rio}.
2) To make things easier in our online format, I have pre-installed all the packages we will be needing on RStudio Cloud.
However, in order to access the functions from these packages, we still need to load them...
Packages
Installing a package puts a copy of it into our personal library that R has access to. In general, we only need to install a package once.
Packages
Installing a package puts a copy of it into our personal library that R has access to. In general, we only need to install a package once.
However, whenever we want to to use a package, we need to load the package in our working session in RStudio.
We load packages with the library() function -- we do this once per session.
Packages
Installing a package puts a copy of it into our personal library that R has access to. In general, we only need to install a package once.
However, whenever we want to to use a package, we need to load the package in our working session in RStudio.
We load packages with the library() function -- we do this once per session.
Loading a package basically makes the contents of that package searchable by R.
In other words, after loading a package, R is able to find the functions included in that package.
You can see what functions are available in your workspace by running the search() function
Your turn 2
03:00
In your RStudio console, look up the help documentation for
import()by typing?import. What do you see?Run
search()in the console. Is the rio package included in this list?Again in the console, load the rio package using the
library()function.Now look again at the help documentation for
import(). What do you see this time?Run
search()again. What is different this time?
Packages
Another package we're going to use a lot going forward is tidyverse.
tidyverse is actually a "meta-package", meaning it contains many individual packages inside of it that are all bundled together.
Packages
Another package we're going to use a lot going forward is tidyverse.
tidyverse is actually a "meta-package", meaning it contains many individual packages inside of it that are all bundled together.
When we load tidyverse we get quite a bit of info.

Packages
Conflicts occur when the same name is used for different things.
Packages
Conflicts occur when the same name is used for different things.
For example, the dplyr package and the stats package (preloaded) both have a function called filter().
Packages
Conflicts occur when the same name is used for different things.
For example, the dplyr package and the stats package (preloaded) both have a function called filter().
When we call filter(), R will only call one of those functions and it might not be the one we want.
Packages
Conflicts occur when the same name is used for different things.
For example, the dplyr package and the stats package (preloaded) both have a function called filter().
When we call filter(), R will only call one of those functions and it might not be the one we want.
Which one will R choose? R has an order in which it searches...
Packages
Conflicts occur when the same name is used for different things.
For example, the dplyr package and the stats package (preloaded) both have a function called filter().
When we call filter(), R will only call one of those functions and it might not be the one we want.
Which one will R choose? R has an order in which it searches...
It starts with the Global Environment, then searches packages in the order that they were loaded, searching more recently loaded packages first.
Packages
Conflicts occur when the same name is used for different things.
For example, the dplyr package and the stats package (preloaded) both have a function called filter().
When we call filter(), R will only call one of those functions and it might not be the one we want.
Which one will R choose? R has an order in which it searches...
It starts with the Global Environment, then searches packages in the order that they were loaded, searching more recently loaded packages first.
You can tell R explicitly that you want a function from a particular package using the notation package::function_name. When in doubt, it's better to use the double colon operator to be specific about which function you want.
Your turn 3
01:00
Look up for the help documentation for
filter()from the stats package.Now look up the help documentation for
filter()from the dplyr package.
Debugging
Before we wrap up, let's talk about error messages.
Debugging
Before we wrap up, let's talk about error messages.
You will run into them constantly, even when using functions you've used many times before -- and especially when using functions/packages that are new to you.
Artwork by @allison_horst
Debugging
We're not going to go into details of debugging, because that could (and should) be a whole course on its own.
But there are a few general things to be aware of...
Debugging
We're not going to go into details of debugging, because that could (and should) be a whole course on its own.
But there are a few general things to be aware of...
Google is your best friend -- it is very likely someone else has had your exact same problem/question before
Some helpful forums are StackOverflow, RStudio Community, CrossValidated
When asking for help, it's best to provide as much context as possible -- best case scenario is to provide a reproducible example
Artwork by @allison_horst
Q & A
05:00